How chimeric reads result from ERV integration. (A to D) A guide to... | Download Scientific Diagram

ChimPipe: Accurate detection of fusion genes and transcription-induced chimeras from RNA-seq data | bioRxiv

Insert size distribution and supported read pairs for data obtained... | Download Scientific Diagram

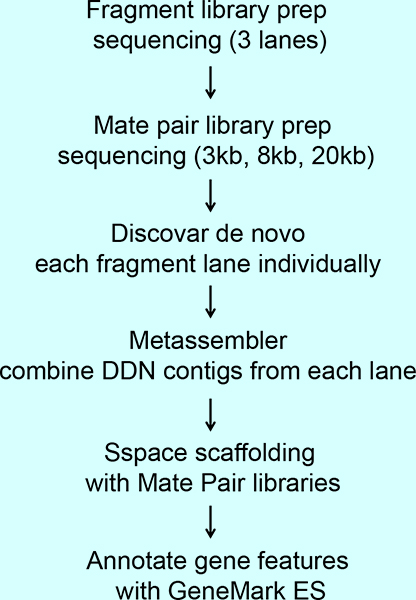
Mate pair sequencing improves detection of genomic abnormalities in acute myeloid leukemia - Aypar - 2019 - European Journal of Haematology - Wiley Online Library
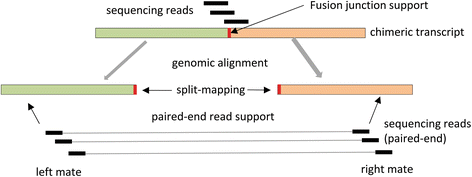
De novo assembly and characterization of breast cancer transcriptomes identifies large numbers of novel fusion-gene transcripts of potential functional significance | BMC Medical Genomics | Full Text

Overlapping Chimeric paired-end alignments are excluded (e.g. in FFPE data) · Issue #285 · alexdobin/STAR · GitHub

Mate-pair signatures for rearrangement style mis-assemblies. (a) Three... | Download Scientific Diagram

HiTea: a computational pipeline to identify non-reference transposable element insertions in Hi-C data | bioRxiv
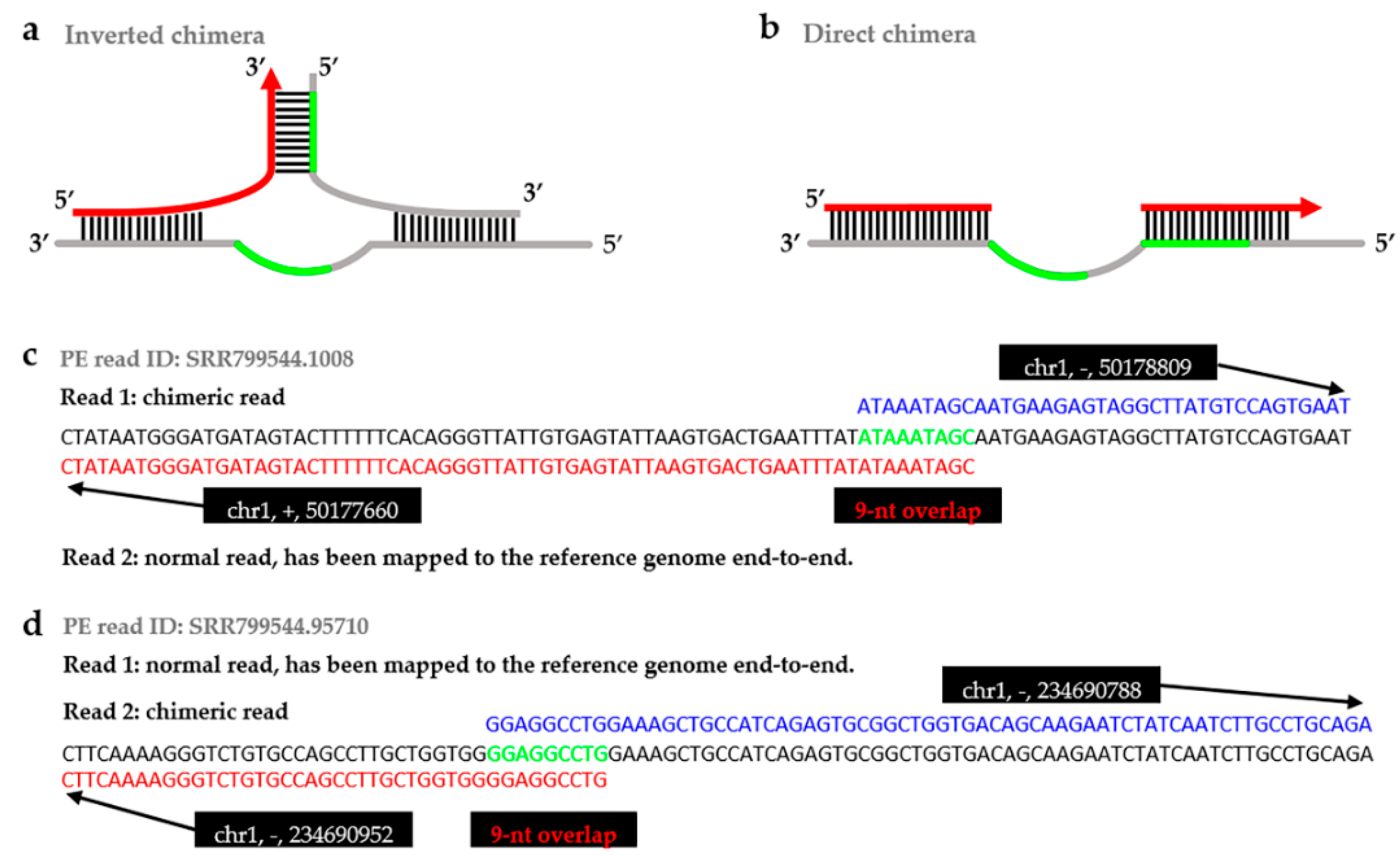
IJMS | Free Full-Text | ChimeraMiner: An Improved Chimeric Read Detection Pipeline and Its Application in Single Cell Sequencing

Mate-pair signatures for inversion style mis-assemblies. (a) Two copy,... | Download Scientific Diagram

Schematic representation of mate-pair library construction by coupling... | Download Scientific Diagram
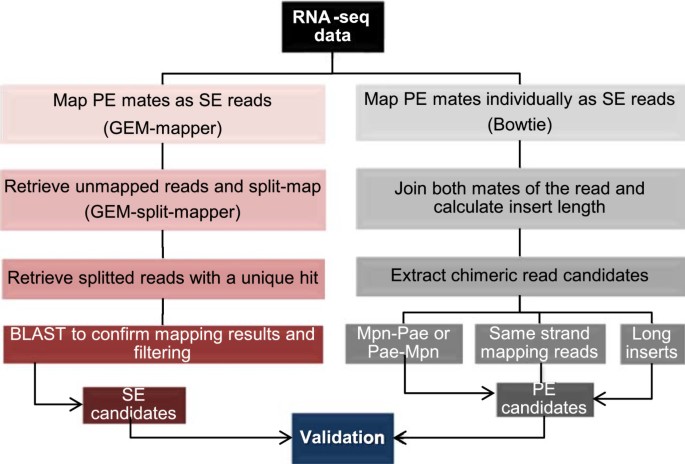
Assessing the hodgepodge of non-mapped reads in bacterial transcriptomes: real or artifactual RNA chimeras? | BMC Genomics | Full Text

Structural variant detection in cancer genomes: computational challenges and perspectives for precision oncology | npj Precision Oncology